ChIPmentation:
fast, cheap, low-input ChIP-seq for histones and transcription factors
Supplementary Website
Overview
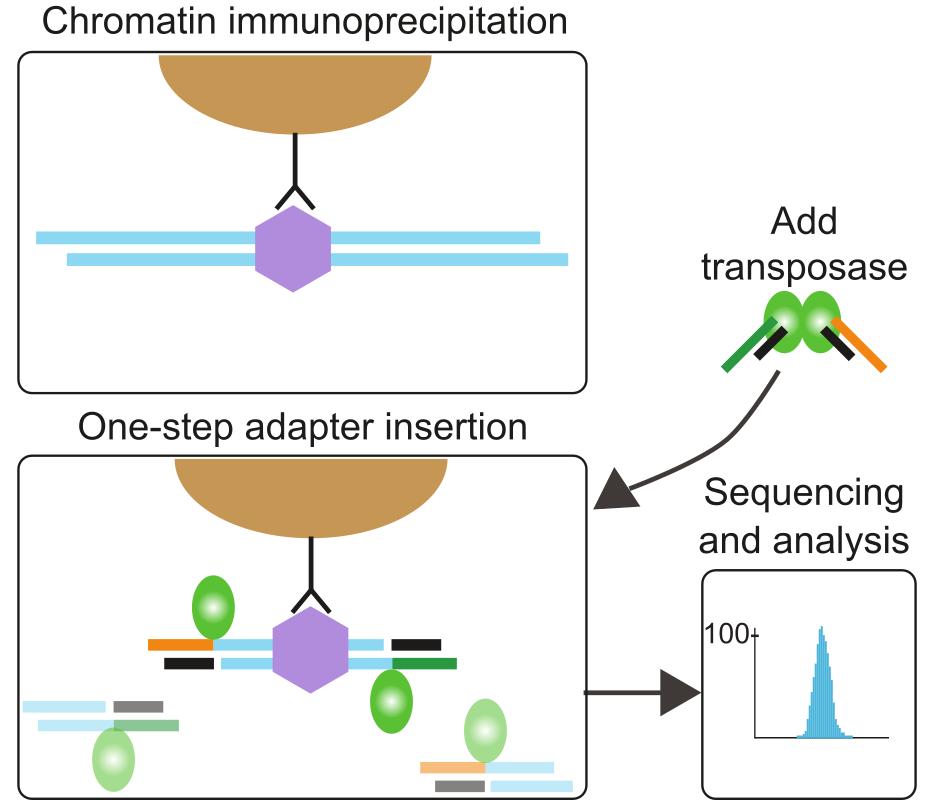
Protocols
This section provides updated ChIPmentation protocols.
| Protocol version | Updated on | Comment | |
| ChIPmentation | v1.14 | 28th September 2016 | Step-by-step protocol. |
| ChIPmentation | v1.1 | 14th October 2015 |
Tested with:
H3K27ac (500k cells), H3K27me3 (500k cells)
CTCF (500k cells), GATA1 (500k cells), PU.1 (500k cells) |
| ChIPmentation | v1.0 (publication) | 10th July 2015 |
Tested with:
H3K4me1 (10M cells), H3K4me3 (10-500k cells), H3K27ac (10M cells), H3K27me3 (10k-500k cells), H3K36me3 (10M cells),
CTCF (100k-10M cells), GATA1 (100k-10M cells), PU.1 (500k-10M cells), REST (100k-10M cells) |
Publications
Here are some examples of publications using ChIPmentation (updated 2018/09/25):
| Title | Journal | DOI | Cell types | Cell number | Antibodies |
| An Integrated and Semiautomated Microscaled Approach to Profile Cis-Regulatory Elements by Histone Modification ChIP-Seq for Large-Scale Epigenetic Studies | Methods Mol Biol | 10.1007/978-1-4939-7896-0_22 | CD4 T cells | 100k | H3K27ac |
| Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks | Nature Communications | 10.1038/ncomms11938 (2016) | CLL cells | 10k-1M | H3K4me1, H3K27ac |
| Epigenome analysis links gene regulatory elements in group 2 innate lymphocytes to asthma susceptibility | J Allergy Clin Immunol | 10.1016/j.jaci.2017.12.1006 | ILC2 | NA | H3K4me2 |
| Genome-wide CRISPR screens in T helper cells reveal pervasive cross-talk between activation and differentiation | bioRxiv | 10.1101/196022 | T cells | 1M | IRF4 (sc-6059), BATF (sc-100974) and FLAG (Sigma M2, #F3165) |
| High-Throughput ChIPmentation: freely scalable, single day ChIPseq data generation from very low cell-numbers | BioRxiv | 10.1101/426957 | MEC1 cells | 100 to 150k | H3K27ac, CTCF |
| HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. | Cancer Discovery | 10.1158/2159-8290.CD-17-0583 | ALL-SIL cells | 20M-40M | H3K27me3 (Millipore 07-449 or Active Motif 39155); H3K4me3 (Active Motif 39159); H3K4me1 (Active Motif 39297); H3K27ac (4729 or Active Motif 39134); STAT5 (Cell Signaling 8363); HOXA9 (Sigma HPA061982) |
| Integrative multi-omics analysis of intestinal organoid differentiation | Mol Sys Biol | 10.15252/msb.20188227 | Intestinal crypt–villus structure derived organoid culture | 80k-150k | Hnf4g, H3K27me3, H3K4me3, and H3K27ac |
| Mutant NPM1 Maintains the Leukemic State through HOX Expression | Cancer Cell | 10.1016/j.ccell.2018.08.005 | OCI-AML3 | NA | H3K27me3, H3K4me3, and H3K27ac |
| RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes | Plos Genetics | 10.1371/journal.pgen.1007233 | Spermatocytes and spermatids | NA | H3K4me2, H3K27ac |
| Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation | Cell | 10.1016/j.cell.2017.02.021 | ILCs | 20-50k | H3K4me2 |
| The Helicase Aquarius/EMB-4 Is Required to Overcome Intronic Barriers to Allow Nuclear RNAi Pathways to Heritably Silence Transcription | Developmental Cell | 10.1016/j.devcel.2017.07.002 | C. elegans | 50k | H3K9me3 |
| The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes | Genes Development | 10.1101/gad.307769.117 | Limb tissue | NA | CTCF |
| The role of TCF3 as potential master regulator in blastemal Wilms tumors | Int J Cancer | 10.1002/ijc.31834 | Xenograft Wilms tumor cell cultures | NA | H3K4me3, and H3K27ac |
| The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes | Nature Genetics | 10.1038/s41588-018-0140-x | Third-instar larvae | NA | GFP |
| Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate | Cell Stem Cell | 10.1016/j.stem.2018.08.016 | B cells reprogrammed to iPS cells | 100k | H3K4me2 |
| Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming | Nature Genetics | 10.1038/s41588-017-0030-7 | Reprogrammed B cells | 100k | H3K4me2 |
| Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation | Nature Immunology | 10.1038/ni.3818 | Macrophages | NA | IRF1 |
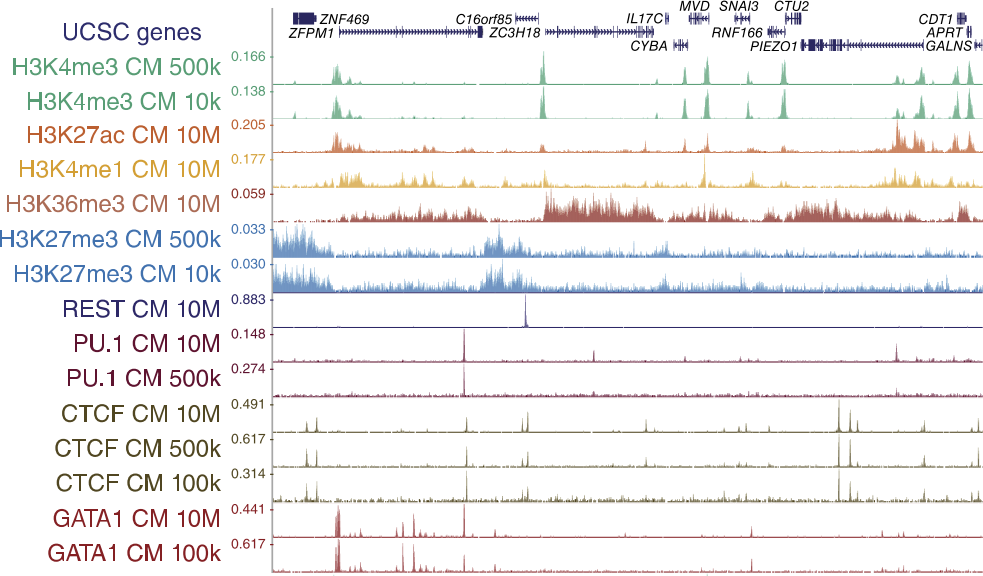
Genome browser tracks
52 libraries were sequenced for ChIPmentation, 24 libraries for standard ChIP-seq, nine libraries for ChIP-tagmentation, and two libraries for ATAC-seq.
Data
52 libraries were sequenced for ChIPmentation, 24 libraries for standard ChIP-seq, nine libraries for ChIP-tagmentation, and two libraries for ATAC-seq.
Software
To foster reproducibility and facilitate reuse, the source code underlying the analysis is contained in a Git repository at Github.
Citation
If you use these data in your research, please cite:
Christian Schmidl*, André F. Rendeiro*, Nathan C. Sheffield and Christoph Bock. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nature Methods 12, 963–965 (2015), doi:10.1038/nmeth.3542
* Shared first authorsNo journal subscription? Ask us to send you the PDF.